Login NTU Bio-Tech
Login NTU Bio-Tech
| Image | Category | Subject | Research Direction | Professor |
|---|---|---|---|---|

|
Research Lab | Lab of Epigenetics and Development | Using mammalian and chicken germ cells and stem cells as materials, study the role of epigenetic regulation in cell fate determination | Shau-Ping Lin |

|
Research Lab | 花卉生技實驗室 | 植物與植物菌質體交互作用、植物型態發育的決定、微藻基因體學研究 | Jen-Chih Chen |

|
Research Lab | Lab of Stem Cell and Regenerative Medicine | 以幹細胞建立藥物毒性檢驗平台、蠑螈肢體再生的基因體學和生物學 | Hsuan-Shu Lee |

|
Research Lab | 微陣列實驗室 | 利用高通量晶片技術探討輻射線抑制腫瘤生長之調節機制、開發高通量定序技術相關之演算法 | Mong-Hsun Tsai |

|
Research Lab | Lab of Reproductive Biotechnology | Nuclear reprogramming by somatic cell nuclear transfer, pre-implantation stages of mammalian embryo development and embryonic stem cell biology | Li-Ying Sung |

|
Research Lab | Lab of Applied and Food Microbiology | 1. Development and application of biomaterials. 2. Bioenergy from agricultural wastes 3. Cell/enzyme immobilization technology 4. Development of functional food | Kuan-Chen Cheng |

|
Research Lab | Lab for Microbial Biotechnology | protein structural biology,expression of probiotic expression system,mycotoxin-detoxifying agent | Je-Ruei Liu |
|
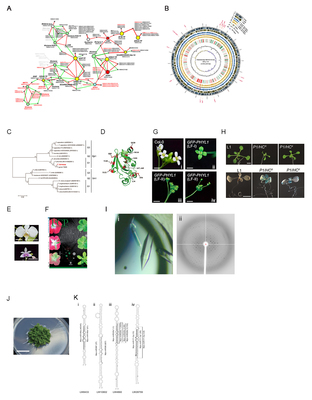
Research Lab | Laboratory of Plant Molecular Biology & Virology | There are existed many difference interactions between microorganisms and plants. The pathogen and probiotic were defined from the microorganisms that based on their behaviors on the host plants. The plant diseases are caused by pathogens, whereas probiotics trigger the plant defense and help the plant growth. One of research interests in our Lab is the mechanisms of interaction between pathogen/probiotics and plants. To address this research interest, the various approaches, such as big-data of bioinformatics, microbiology, biotechnology, and molecular biology, were used to study the mechanisms of pathology/resistance. These findings provide information for our lab to innovate a new drug for plant disease or novel biologics for promoting plant growth and defense responses. Research subjects in our Lab (1) Big-data of bioinformation We used next-generation sequencing (NGS) approach to study the genome projects of microorganisms and plants. Several supercomputing infrastructures from National Taiwan University (NTU) and National Center for High-Performance Computing (NCHC) support our lab for big-data mining to understand the mechanism of defense or disease through the gene-to-gene network relationship between microorganism and plants. For example, phytoplasma triggers leafy flower formation on infected plants. Through the big-data mining, we identified the mechanism of the leafy flower. We also found that the amount of vinblastine (an anti-cancer drug) were increased in phytoplasma infected plants. For applying this finding on the innovation of new anti-cancer drug, our lab is studying the mechanism through combining approaches of the big-data mining and biotechnology (Fig. 1A). We also focus on the genome study of importance microorganism. For example, we cooperate with Prof. Yuan-Tay Shyu (Department of Horticulture & Landscape Architecture) to solve the whole genome sequence of halotolerant bacteria (original of Taiwan) and identified several critical genes that involve in halotolerant (Fig. 1B). The information will be applied in biotechnology for developing the salt-resistance enzymes. (2) Development of biologics We identified Epl1 effector from Trichoderma spp., which can trigger plant innate immunity (Fig. 1C). We purified the recombinant protein of Epl1 that has trigger plant defense ability on various pathogen diseases. Moreover, we also study the gene-to-gene network on Epl1-mediated defense through the transcriptome of NGS for modification of Epl1 to develop new a biologic for plant protection (Fig. 1D). (3) Metabolomics Instead of the gene-to-gene network, we also study the metabolite profiles under pathogen infected plants. For instance, the pigment biosynthesis in the petals of an orchid flower and vinblastine biosynthesis on the phytoplasma infected plants (Fig. 1E & F) We are studying how to apply these finding to increase these valuable phenotypes through the biotechnology approach. (4) Studying the mechanism of effector suppresses gene silencing pathway We also investigate that how effectors of pathogens interfere gene silencing pathway. The PHYL1 effector of phytoplasma inhibits miR396 expression, resulting in miss-regulation of miR396 on SVP target mRNA. The SVP is an important negative flowering regulator; therefore, the miss-regulation on miR396-SVP mRNA causes leafy flower formation (Fig. 1G). In addition, the P1/HC-Pro of potyvirus is a gene silencing suppressor that inhibits miRNA pathway, resulting in symptoms development (Fig. 1H). Through modification of P1/HC-Pro gene can generate mild strain virus, which shown symptomless on the infected plant that triggers cross-protection to against severe strain virus infection. Instead of studying the mechanism of cross-protection, we also investigate the mechanism of P1/HC-Pro suppression ability. Furthermore, we have developed the mild strain as a viral vector for biotechnology application. (5) Protein engineering In our lab, we focus on the recombinant protein of construction and purification for effectors or critical enzymes of plant. Through various large scale purification systems, we purify recombinant proteins for studying in the x-ray crystallography and biochemistry activity. These recombinant proteins can be applied in enzyme engineering, biologics, and protein drug developments (Fig. 1I). International cooperation (1) International cooperation on Marchantia polymorpha liverwort Our lab joined the international society of Marchantia polymorpha liverwort to study the identification and annotation of miRNA genes and their targets (Fig. 1J & K). (2) International cooperation Our lab welcomes international students visiting and cooperation. We have many cooperators around the world, including in UK, Spain, Japan, Australia, France, Mexico, and Germany. Every year, our students have a chance to visit these labs for short stay on their research projects. We have also International exchanged students in our lab for their internship. | Shih-Shun Lin |

|
Research Lab | Lab of Microbial Ecology & Functional Engineering | Chi-Te Liu is head of Microbial Ecology & Functional Engineering Lab, and also a faculty at Institute of Biotechnology in National Taiwan University. He studied environmental microbiology and received his doctor degree in Applied Biological Chemistry at Graduate School of Agricultural and Life in The University of Tokyo. He worked on plant-microbe symbiosis when he was a post-doc at Biotechnology Research Center in The University of Tokyo. His research expertise is on Agricultural Biotechnology, Environmental Microbiology and Microbe-Plant interactions (Symbiosis). His current research projects are as follows: 1. Development of multifunctional PGPR agents [free-living nitrogen-fixing bacteria [Azorhizobium caulinodans], phototropic bacteria (Rhodopseudomonas palustris), biocontrol bacteria (Bacillus amyloliquefaciens), etc] and their applications in fields and plant factory. 2. Exploring the molecular mechanisms of plant-microbe interactions, especially those for plant growth promotion traits and biological nitrogen fixation. 3. Characterization of the roles of Type VI secretion system in A. caulinodans under vegetative and symbiotic conditions. | Chi-Te LIU |

|
Research Lab | Lab of Immuno-Oncology | Dr. Shu-Han Yu’s laboratory has a well-defined focus on the immune mechanisms in the tumor microenvironment, adoptive cell therapy, and the potential role of the microbiome in cancer initiation and progression. | Shu-Han Yu |